Folding of protein into its unique biologically active state is an intricate process. Protein refolding under in vitro conditions may be encouraged by either stabilizing native state of protein or via accelerating the kinetics that leads to precise folding reaction. Additionally, overpowering undefined aggregation of the unfolded and/or intermediates on the folding pathway can also aid in refolding protein structure. The protein folding process becomes even more complex in case of multidomain proteins (such as albumins), where inter-domain interactions can affect the complete folding process. Bovine serum albumin (BSA) and human serum albumin (HSA) are examples of such multidomain proteins. BSA and HSA are globular proteins, they function as a biological carrier of fatty acid anions and other simple amphiphiles in the bloodstream. BSA and HSA are highly significant proteins from a biopharmaceutical perspective, as they are models for studying drug-protein interaction in vitro. Numerous experiments with the aim of characterizing the binding capacity and binding sites of albumins have been carried out.
Over the past two decades, various studies have established in ionic liquid (IL) as a potential biocompatible solvent for proteins, particularly on enzyme-catalyzed reactions. One of the major challenges in protein stability is that proteins can easily unfold in the presence of denaturants like urea, which alters the native structure of proteins. There are numerous studies in which ionic liquids (ILs) act as promising biocompatible solvents (Bio-IL) for biomolecules. In this context, we present the refolding ability of a biocompatible imidazolium-based ionic liquid, 1-ethyl 3-methyl imidazolium ethyl sulfate [Emim][ESO4] (IL-1) and 1-ethyl 3-methyl imidazolium chloride [Emim][Cl] (IL-2) against the chemically induced structural changes in bovine and human serum albumin (BSA and HSA). The work is substantiated with several spectroscopic, thermal and docking studies. In steady-state fluorescence spectroscopy, we observe that the emission intensity quenches for the protein in urea which is reversible with the addition of ILs. Circular dichroism spectral studies reflect the reappearance of α helical content which is a good indicator of the refolding ability of ILs. Further, thermal fluorescence studies showed that ILs has the ability to refold the urea-induced denatured protein at higher temperature range only up to 7 M urea concentration, however, above 7 M urea concentration IL somehow fails to refold the protein. The work is also supported by dynamic light scattering (DLS) measurements, the degree of BSA/HSA aggregation was reduced with the introduction of Bio-IL to the urea-BSA/ urea-HSA system ensuring the aggregate-free refolding. Furthermore, molecular docking studies were employed to probe the binding sites and the results are well corroborated with the spectroscopic and thermal folding results. Therefore, through this paper, we aim to unravel the mechanistic intricacy of ILs using experimental and docking approaches. Overall, ILs act as recoiling medium for both native and unfolded (denatured by urea) of BSA/HSA native structure (Figure).
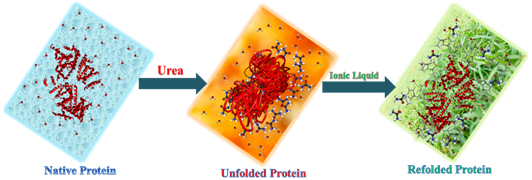
Ref: Anamika Sindhu, Kavya Bhakuni, Kamatchi Sankaranarayanan*, Pannuru Venkatesu*. Implications of Imidazolium based Ionic Liquids as Refolding Additives for Urea-induced denatured Serum Albumins. ACS Sustainable Chemistry & Engineering, 2020, Vol.8, 604. DOI: 10.1021/acssuschemeng.9b06194







